Tuesday, September 24, 2024
Unlocking the Future of Medicine: A Deep Dive into Gene Editing


Imagine a world where genetic diseases like cystic fibrosis, HIV, or even certain cancers could be cured by editing the very DNA that causes them. Welcome to the fascinating realm of gene editing — a groundbreaking frontier in modern science that’s reshaping our understanding of life itself. In this blog, we explore the powerful gene-editing tools CRISPR-Cas9, TALENs, and Zinc Finger Nucleases (ZFNs), each offering remarkable precision to rewrite the genetic code.
From correcting faulty genes in mice to the potential of editing human embryos, these technologies hold the promise of transforming medicine as we know it. But with great power comes great responsibility — what are the ethical considerations, and how do we navigate the potential risks, such as unintended genetic modifications? We’ll also dive into real-world applications, from curing genetic disorders to tackling diseases like HIV, offering a glimpse into how these innovations could redefine the future of healthcare.
Whether you’re a science enthusiast, a student, or just curious about the next big leap in biotechnology, this blog will guide you through the exciting, and sometimes controversial, landscape of gene editing. Get ready to uncover the science, the possibilities, and the future that lies ahead.
CRISPR-CAS9:
Clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 is a gene-editing technology causing a major upheaval in biomedical research. It makes it possible to correct defects in the genome. It has already been demonstrated that it can be used to repair the defects in the DNA of mice curing them of genetic disorders ,and it has been reported that human embryos can be similarly modified. It also has a number of laboratory applications including rapid generation of cellular and animal models, functional genomic screens ,live imaging of the cellular genome,gene therapy ,treating HIV and other diseases.
It has two components:
- Guide RNAs to take the Cas9 to the desired target gene.
- Cas9(CRISPR-associated protein 9)-the endonuclease which breaks a double stranded DNA and hence modifying genome
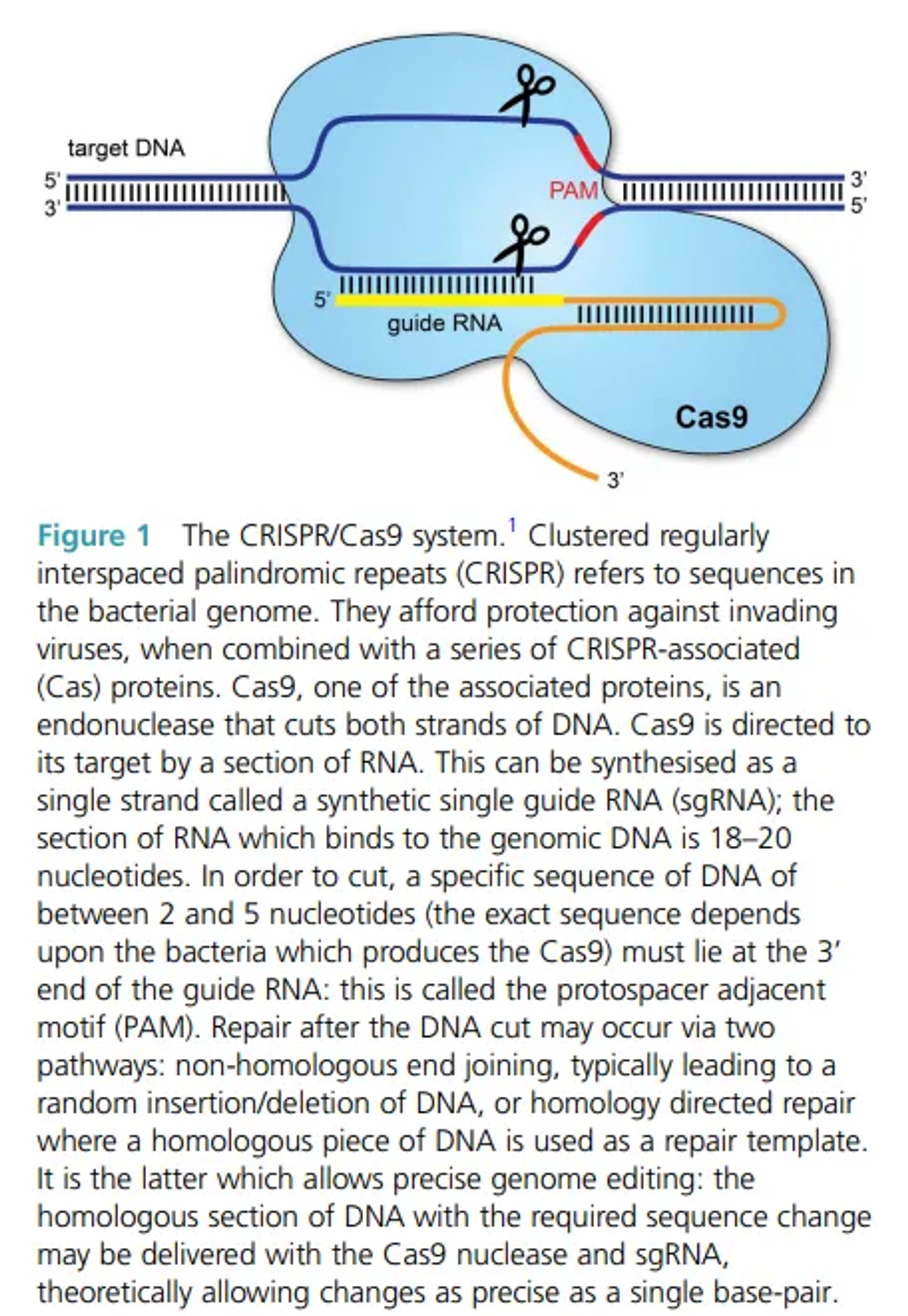
Application:
- Correction of genetic disorders such as cystic fibrosis (CF), Duchenne’s muscular dystrophy (DMD) and haemoglobinopathies found in humans and mice(DMD).The approach so far has currently only been validated in preclinical models, but there is hope it can soon be translated to clinical practice.
- Treatment in HIV
- Engineering somatic cells ex vivo to treat malignancy or other diseases
Limitations:
- To safely deliver Cas9-nuclease encoding genes and guide them to the right cells. Guide RNAs such as AAV may be too small to allow efficient transduction of the Cas9 gene.
- To counter the above reason a smaller Cas9 gene could be used, but this has additional implications on efficacy.
- Another significant concern is the possibility of off target effects on parts of the genome separate from the region being targeted. Unintentional edits of the genome could have profound long-term complications for patients, including malignancy.
- The concentration of the Cas9 nuclease enzyme and the length of time Cas9 is expressed are both important when limiting off-target activity.
TALENs (Transcription Activator-Like Effector Nucleases):
TALENs are a type of engineered nuclease used for genome editing. They consist of a DNA-binding domain derived from transcription activator-like effectors (TALEs) and a nuclease domain from FokI.
Components
DNA-Binding Domain (TALEs)
- TALE proteins are derived from Xanthomonas bacteria.
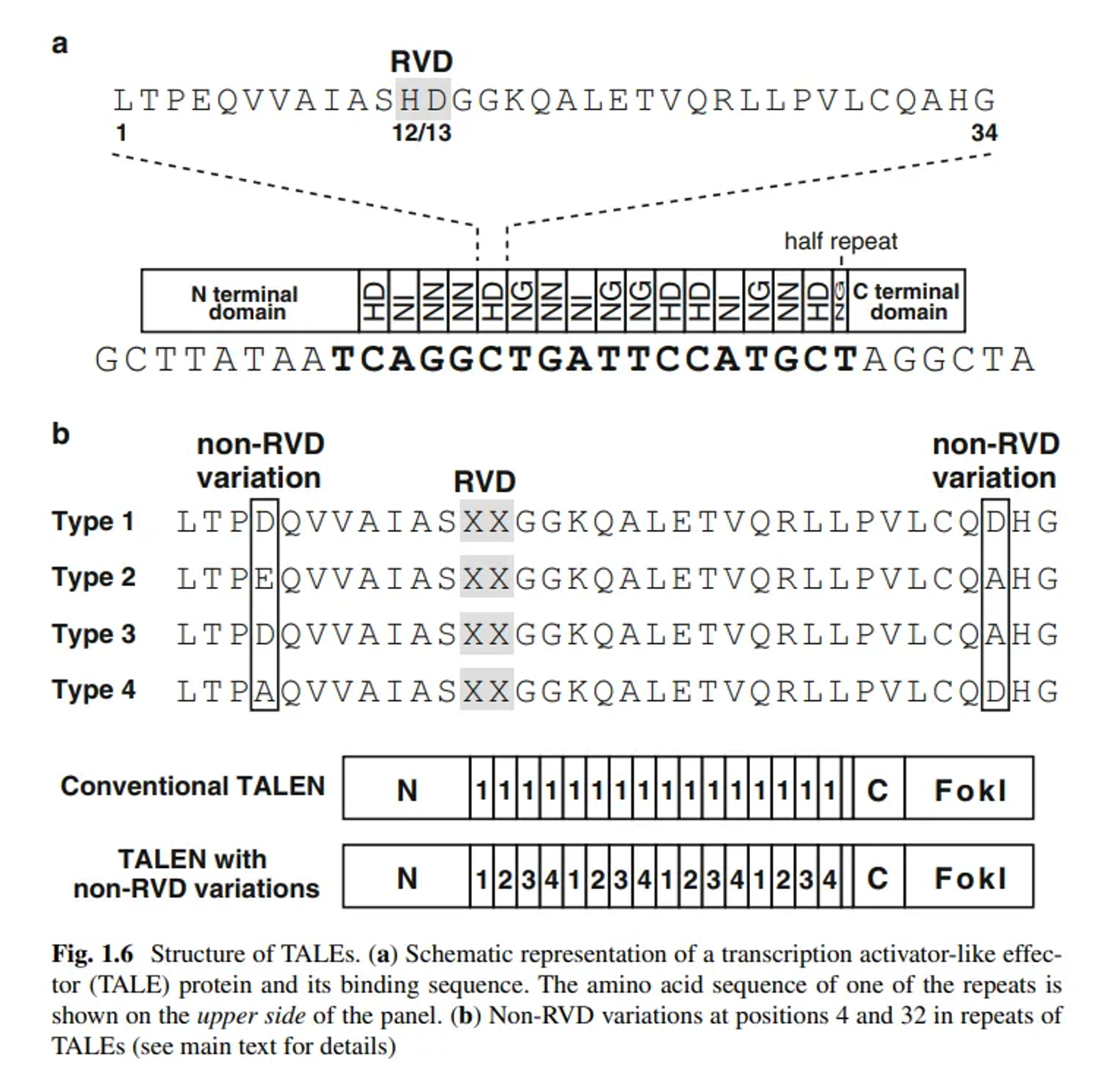
- They contain tandem repeats, typically 33–35 amino acids long, where each repeat recognizes a single DNA base
- Each repeat has a repeat-variable diresidue (RVD) that determines the DNA base specificity.
Nuclease Domain (FokI)
- The FokI nuclease domain is a non-specific DNA cleavage domain.
- It requires dimerization to cut DNA, thus two TALEN monomers bind to opposite strands of the target DNA, bringing the FokI domains together to cleave.
Step-by-Step Working of TALENs
1. Design and Construction
DNA Binding Domain Design
TALE Proteins Origin:
- TALE (Transcription Activator-Like Effector) proteins are derived from Xanthomonas bacteria. These bacteria use TALEs to alter the expression of genes in host plants, facilitating infection.
Structure of TALE Repeats:
- Each TALE repeat is a protein module of 33–35 amino acids. The sequence is highly conserved, except for two positions known as the repeat-variable diresidue (RVD).
- The RVDs at positions 12 and 13 are crucial because they determine the specific DNA base that each repeat will recognize:
- NI recognizes adenine (A).
- HD recognizes cytosine ©.
- NG recognizes thymine (T).
- NN recognizes guanine (G) (and sometimes adenine (A)).
- By linking multiple TALE repeats in a specific order, a custom DNA-binding domain is designed to target a specific DNA sequence.
Fusion with FokI Nuclease Domain
FokI Nuclease:
- The FokI nuclease is a bacterial enzyme that cleaves DNA. However, its cleavage domain requires dimerization to function, meaning it needs to pair with another FokI domain to cut DNA.
- In TALENs, the DNA-binding domain (customized TALE repeats) is fused to the FokI nuclease domain.
- For the FokI domains to dimerize and cut the DNA, two TALEN monomers must bind to adjacent sites on opposite DNA strands. This ensures that the cleavage occurs at a specific, targeted location.
2. Assembly and Delivery
Vector Construction
Cloning into Vectors:
- The TALEN constructs, which include the custom TALE repeats and the FokI nuclease domain, are cloned into appropriate expression vectors. These vectors can be plasmids, viral vectors, or other delivery systems.
- The vector ensures that the TALENs are expressed efficiently within the target cells.
Introduction into Target Cells
Delivery Methods:
- Electroporation: An electrical field is applied to cells to increase the permeability of the cell membrane, allowing the vectors to enter.
- Microinjection: The vectors are directly injected into the cells using a fine needle.
- Viral Transduction: Modified viruses are used to deliver the TALEN constructs into the cells. This method can be very efficient, especially for difficult-to-transfect cells.
3. Binding to Target DNA
TALEN Monomer Binding
Specific DNA Binding:
- Inside the cell, each TALEN monomer binds to its specific DNA sequence through the TALE repeats.
- One TALEN monomer binds to a specific sequence on one strand of the DNA, and the other monomer binds to an adjacent sequence on the opposite strand.
- This specific binding positions the FokI nuclease domains in close proximity, ready to dimerize.
4. DNA Cleavage
Dimerization of FokI Nuclease
Activation of Cleavage:
- When the two TALEN monomers are correctly bound to the target DNA sequences, the FokI nuclease domains dimerize.
- Dimerization activates the nuclease activity of FokI, leading to the creation of a double-strand break (DSB) at the specific target site in the genome.
5. Cellular Repair Mechanisms
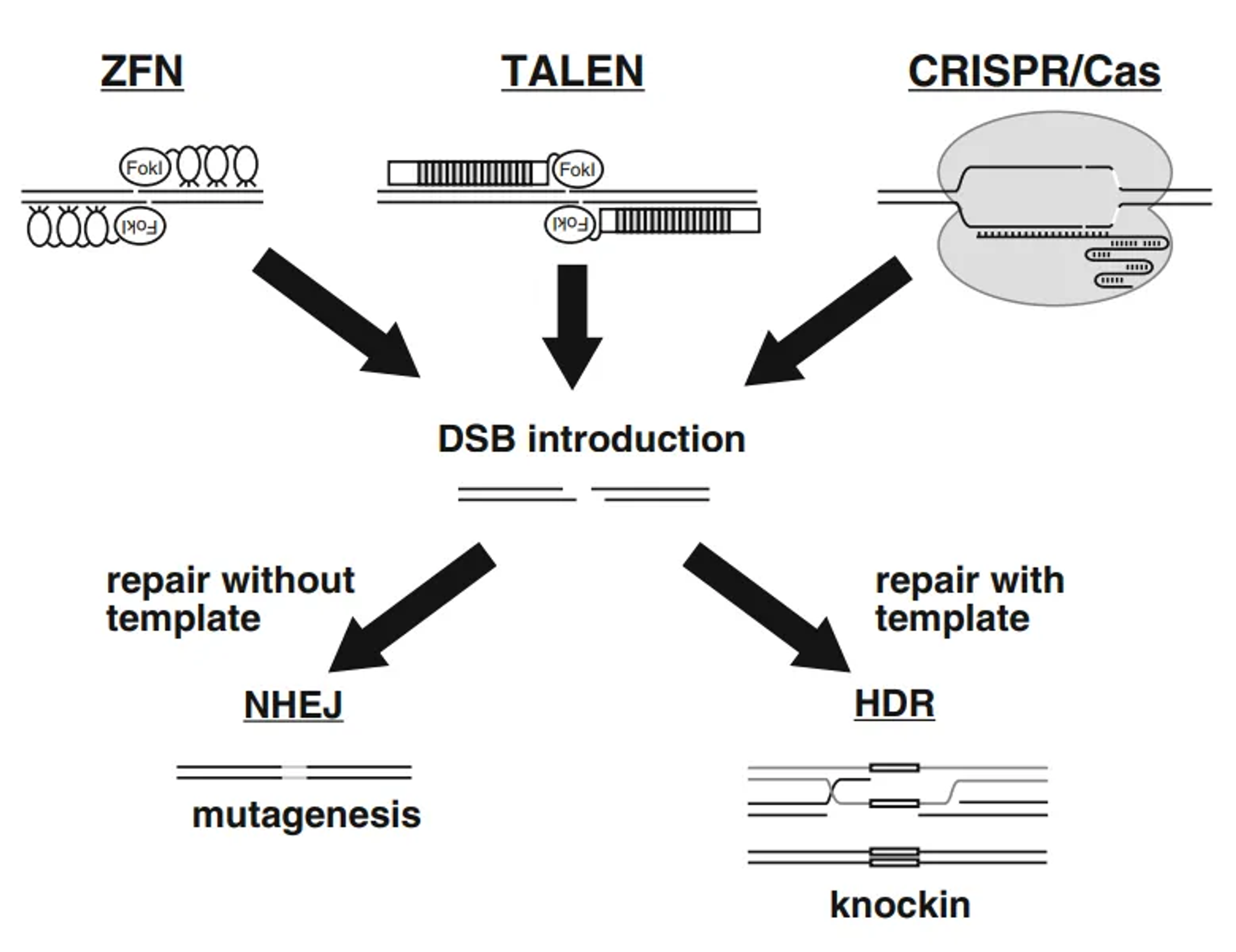
Non-Homologous End Joining (NHEJ)
Quick Repair Process:
- The cell’s natural repair machinery detects the DSB.
- NHEJ is the primary repair mechanism for DSBs and works by directly ligating the broken DNA ends together.
- Error-Prone Nature: NHEJ is quick but error-prone. It often results in small insertions or deletions (indels) at the break site. These indels can disrupt the coding sequence of genes, potentially leading to loss-of-function mutations. This property is useful for gene knockout experiments.
Homology-Directed Repair (HDR)
Accurate Repair Process:
- If a donor DNA template with sequences homologous to the regions flanking the DSB is provided, the cell can use HDR to repair the break.
- Using the Donor Template: HDR uses the donor template to guide the repair, allowing for precise insertion or correction of sequences at the break site. This method is used for gene correction or insertion of new genetic material.
- Precision: HDR is less error-prone than NHEJ and allows for precise genetic modifications. It’s particularly useful for therapeutic applications where specific mutations need to be corrected or new genes need to be inserted.
Applications of TALENs
Gene Knockout:
- By introducing indels via NHEJ, TALENs can disrupt gene function, creating knockout models for studying gene function or disease mechanisms.
Gene Correction:
- Using HDR, TALENs can correct mutations in genes, potentially providing therapeutic benefits for genetic disorders.
Transgenic Organisms:
- TALENs facilitate the insertion of new genes or regulatory elements into the genomes of model organisms, aiding in research and biotechnology.
Functional Genomics:
- TALENs are used to study gene function and regulation by creating targeted modifications and analyzing their effects on cellular processes.
ZFNs (Zinc Finger Nucleases)
Zinc Finger Nucleases (ZFNs) are engineered proteins that combine the DNA-binding specificity of zinc finger domains with the DNA-cleaving capability of the FokI nuclease domain. They are used to create targeted double-strand breaks (DSBs) in the genome, which the cell then repairs, leading to specific genetic modifications.
Components:
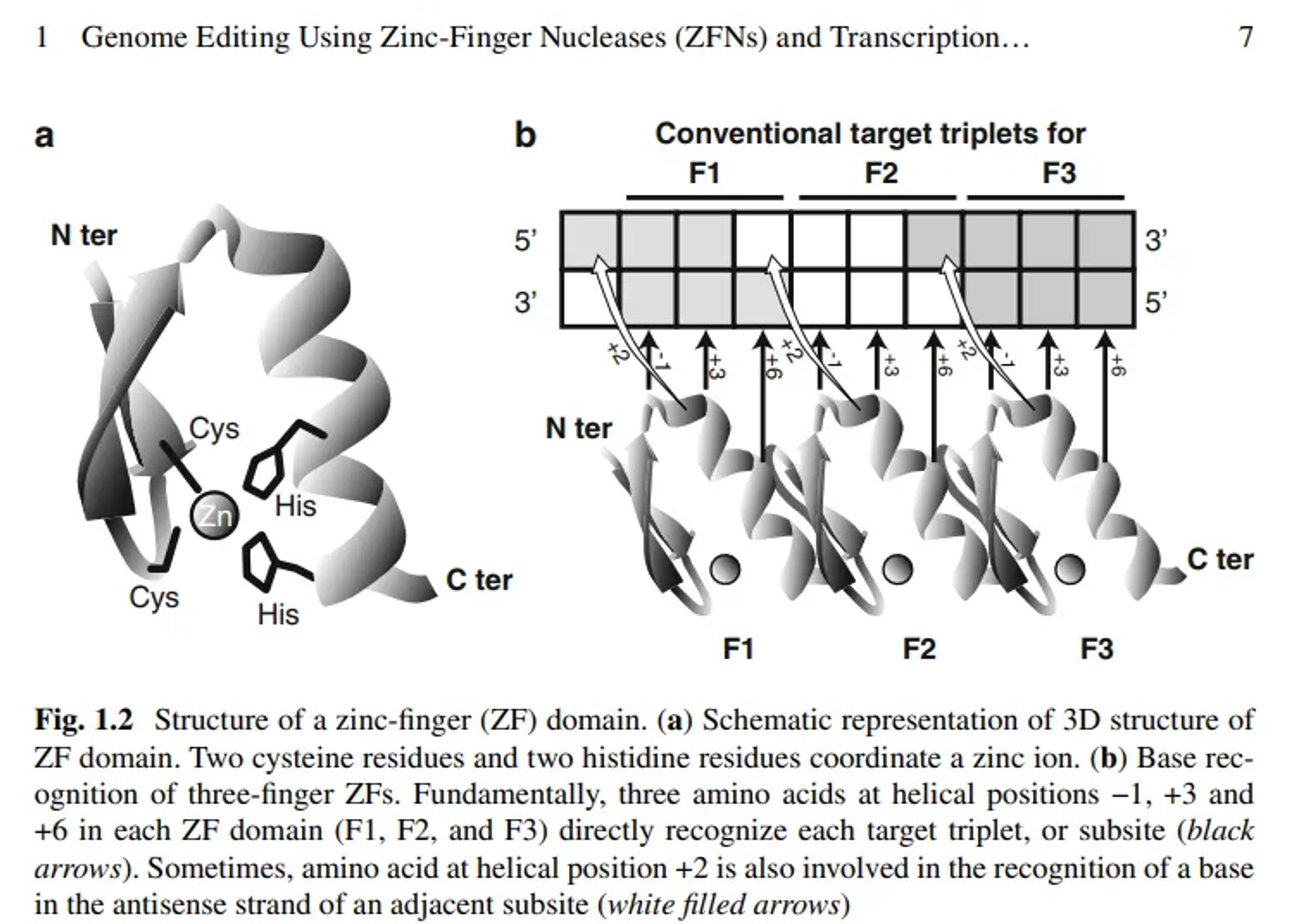
DNA-Binding Domain (Zinc Fingers):
- Zinc fingers are small protein motifs that can bind to specific DNA sequences.
- Each zinc finger typically recognizes 3–4 base pairs of DNA.
- Multiple zinc fingers can be linked together to recognize longer DNA sequences.
Nuclease Domain (FokI):
- As with TALENs, the FokI nuclease domain in ZFNs requires dimerization to cut DNA.
Step-by-Step Working of ZFNs
1. Design and Construction
Zinc Finger DNA-Binding Domain Design
Zinc Finger Structure:
- Zinc fingers are small protein motifs stabilised by a zinc ion. Each zinc finger domain typically consists of about 30 amino acids that form a ββα structure.
- Each zinc finger recognizes a specific 3-base pair (bp) DNA sequence.
Recognition Code:
- The amino acids at positions -1, 2, 3, and 6 of the α-helix within each zinc finger interact with the DNA bases.
- By customizing these positions, scientists can engineer zinc fingers to bind specific DNA triplets.
Modular Assembly:
- Multiple zinc fingers can be linked together in tandem to recognize longer DNA sequences. For instance, combining 3 zinc fingers allows for recognition of a 9-bp sequence, while combining 6 zinc fingers can recognize an 18-bp sequence.
- The longer the sequence, the higher the specificity and the lower the likelihood of off-target effects.
Optimization:
- The sequence and structure of each zinc finger domain are optimized to ensure strong and specific binding to the target DNA sequence.
- Computational tools and experimental validation help in designing effective zinc finger arrays.
Fusion with FokI Nuclease Domain
FokI Nuclease:
- The FokI nuclease is a bacterial enzyme that cleaves DNA. Its nuclease domain is non-specific and requires dimerization to cut DNA.
- The DNA-binding domain of FokI is replaced with custom zinc fingers to target specific DNA sequences.
Monomer Fusion:
- Each ZFN monomer consists of several zinc fingers fused to the FokI nuclease domain.
- To ensure specificity, two ZFN monomers must bind to adjacent DNA sequences on opposite strands, positioning the FokI domains to dimerize and cleave the DNA.
2. Assembly and Delivery
Vector Construction
Cloning into Vectors:
- The designed ZFN constructs are inserted into expression vectors, such as plasmids or viral vectors.
- These vectors contain promoter sequences to drive the expression of ZFNs in the target cells.
Expression Optimization:
- The vectors are engineered to optimise the expression levels of ZFNs, ensuring efficient binding and cleavage.
- Regulatory elements, such as enhancers and terminators, may be included to enhance expression.
Introduction into Target Cells
Delivery Methods:
- Electroporation: An electrical field temporarily opens pores in the cell membrane, allowing the vectors to enter the cells.
- Microinjection: Vectors are directly injected into cells using a fine needle, providing precise delivery.
- Viral Transduction: Modified viruses, such as lentiviruses or adenoviruses, deliver ZFN constructs into cells. This method is efficient for hard-to-transfect cells.
Selection and Validation:
- After delivery, cells expressing ZFNs are selected and validated using molecular biology techniques, such as PCR and sequencing.
3. Binding to Target DNA
ZFN Monomer Binding
Specific DNA Binding:
- Inside the cell, each ZFN monomer binds to its specific DNA sequence through the zinc finger domains.
- The zinc finger domains recognize and bind to their target DNA triplets with high specificity.
Target Sequence:
- One ZFN monomer binds to a sequence on one DNA strand, and the other monomer binds to an adjacent sequence on the opposite strand.
- The optimal distance between binding sites is usually 5–7 bp, allowing proper dimerization of the FokI nuclease domains.
4. DNA Cleavage
Dimerization of FokI Nuclease
Activation of Cleavage:
- When the two ZFN monomers bind to their target sequences, the FokI nuclease domains are brought into close proximity.
- The FokI domains dimerize, activating the nuclease activity.
Double-Strand Break (DSB):
- The dimerized FokI nuclease cuts both DNA strands at the target site, creating a DSB.
- The DSB is a critical trigger for the cell’s DNA repair mechanisms.
5. Cellular Repair Mechanisms
Non-Homologous End Joining (NHEJ)
Quick Repair Process:
- The cell’s repair machinery detects the DSB and activates the NHEJ pathway.
- NHEJ ligates the broken DNA ends without needing a homologous template.
Error-Prone Nature:
- NHEJ is fast but error-prone, often resulting in small insertions or deletions (indels) at the break site.
- These indels can disrupt the coding sequence of genes, leading to loss-of-function mutations. This is useful for gene knockout studies.
Homology-Directed Repair (HDR)
Accurate Repair Process:
- If a donor DNA template with sequences homologous to the regions flanking the DSB is provided, the cell can use HDR for repair.
- HDR is a precise repair process that uses the donor template as a guide.
Gene Correction and Insertion:
- HDR allows for the accurate insertion or correction of genetic sequences at the break site.
- This method is ideal for therapeutic applications, such as correcting disease-causing mutations or inserting new genes.
Applications of ZFNs
Gene Knockout: Creating gene knockouts by introducing indels through NHEJ, which is useful for studying gene function and disease mechanisms.
Gene Correction: Using HDR to correct mutations, offering potential therapeutic benefits for genetic disorders.
Transgenic Organisms: Inserting new genes or regulatory elements into the genomes of model organisms, aiding research and biotechnology.
Functional Genomics: Studying gene function and regulation by creating targeted genetic modifications and analyzing their effects on cellular processes.
Comparison of TALENs and ZFNs
Design and Specificity:
- ZFNs: Each zinc finger recognizes 3–4 base pairs, and multiple fingers need to be linked for longer sequences. Designing ZFNs is complex due to context-dependent binding specificities of zinc fingers.
- TALENs: Each repeat recognizes a single base pair, making TALEN design more straightforward and modular. TALENs generally offer higher target specificity and easier customization.
Efficiency:
- Both TALENs and ZFNs can efficiently introduce double-strand breaks at specific genomic locations, but TALENs are often considered more efficient and easier to design for a wide range of sequences.
Off-Target Effects:
- Both technologies can have off-target effects, but TALENs are generally considered to have lower off-target activity due to their longer and more specific DNA recognition sequences.
Technical Challenges:
- ZFNs are more challenging to design and assemble due to the context-dependent nature of zinc finger interactions.
- TALENs, with their modular design, are more straightforward to construct, though they are larger proteins, which can sometimes pose delivery challenges.
Use Cases:
Both are used in gene therapy, functional genomics, and creating genetically modified organisms. TALENs are preferred for applications requiring higher specificity and easier design.
Non-Homologous End Joining (NHEJ)
NHEJ is a quick and relatively error-prone DNA repair mechanism used to fix DSBs by directly ligating the broken DNA ends together without the need for a homologous template.
Step-by-Step Working of NHEJ
1. Detection of DNA Break:
- When a DSB occurs in the DNA, it is immediately recognized by the cell’s damage response machinery.
- The Ku heterodimer, consisting of Ku70 and Ku80 proteins, quickly binds to the broken DNA ends. This binding serves as a scaffold for the assembly of other NHEJ repair proteins and protects the DNA ends from further degradation.
2. Recruitment of DNA-PKcs:
- Once the Ku70/Ku80 heterodimer is bound to the DNA ends, it recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs).
- DNA-PKcs, along with Ku70/Ku80, forms the DNA-PK holoenzyme, which stabilises the DNA ends and initiates further repair processes.
3. Synapsis Formation:
- DNA-PKcs helps to align the broken DNA ends, a process called synapsis. This alignment is crucial for the accurate repair of the break.
4. End Processing:
- The DNA ends often need to be processed to make them compatible for ligation. This processing involves:
- Artemis: A nuclease that trims or resects DNA ends to remove any overhangs or mismatched bases.
- Pol μ and Pol λ: Polymerases that add or fill in missing nucleotides at the DNA ends to create blunt ends or short overhangs that can be easily joined.
5. Ligation of DNA Ends:
- The final step in NHEJ involves the ligation of the processed DNA ends. This step is mediated by:
- DNA Ligase IV: The enzyme responsible for sealing the DNA break.
- XRCC4 and XLF/Cernunnos: These proteins form a complex with DNA Ligase IV, stabilising it and helping to align the DNA ends for efficient ligation.
Detailed Steps in NHEJ:
1. Detection of DNA Break:
Ku70/Ku80 Binding:
- Immediately upon a double-strand break occurring, the Ku70/Ku80 heterodimer binds to the broken DNA ends.
- The Ku proteins form a ring-like structure around the DNA, providing stability and protecting the ends from degradation.
2. Recruitment of DNA-PKcs:
Formation of DNA-PK Complex:
- The DNA-PK complex, composed of DNA-PKcs and Ku70/Ku80, forms at the break site.
- DNA-PKcs is a large protein kinase that phosphorylates various substrates involved in DNA repair and helps to bridge the DNA ends.
3. Synapsis Formation:
Alignment of DNA Ends:
- DNA-PKcs facilitates the bringing together (synapsis) of the two DNA ends, aligning them in preparation for repair.
- This step ensures that the DNA ends are in close proximity and properly oriented for ligation.
4. End Processing:
Artemis-Mediated Processing:
- Artemis is activated by DNA-PKcs and functions to trim overhangs or process complex DNA ends, making them more suitable for ligation.
- Artemis removes any damaged or incompatible nucleotides from the ends.
Pol μ and Pol λ Activity:
- DNA polymerases Pol μ and Pol λ fill in any gaps by adding nucleotides to the ends, ensuring that the DNA ends are compatible for ligation.
- These polymerases are particularly important when the ends are not directly compatible for joining.
5. Ligation of DNA Ends:
XRCC4-Ligase IV Complex:
- The XRCC4 protein binds to DNA Ligase IV, forming a complex that is essential for the final ligation step.
- XRCC4 stabilises Ligase IV and positions it correctly at the DNA ends.
Role of XLF/Cernunnos:
- XLF (also known as Cernunnos) interacts with XRCC4 and enhances the activity of the Ligase IV-XRCC4 complex.
- This interaction is crucial for efficient ligation, especially when there are small gaps or misalignments at the DNA ends.
Final Ligation:
- DNA Ligase IV catalyses the formation of phosphodiester bonds between the DNA ends, sealing the break and restoring the integrity of the DNA molecule.
- This step completes the NHEJ repair process.
Importance and Characteristics of NHEJ:
Error-Prone Nature:
- NHEJ is relatively quick and efficient but can introduce errors at the repair site. Small insertions or deletions (indels) often occur because the process does not rely on a template.
- These indels can disrupt the coding sequence of genes, potentially leading to loss-of-function mutations.
Cell Cycle Independence:
- NHEJ is active throughout the cell cycle, including in non-dividing cells, making it a versatile repair mechanism for various cell types and conditions.
Role in Genome Editing:
- NHEJ is exploited in genome editing techniques, such as CRISPR-Cas9, where inducing a DSB can lead to gene disruption through the error-prone repair process.
- This property is useful for creating gene knockouts or studying gene function.
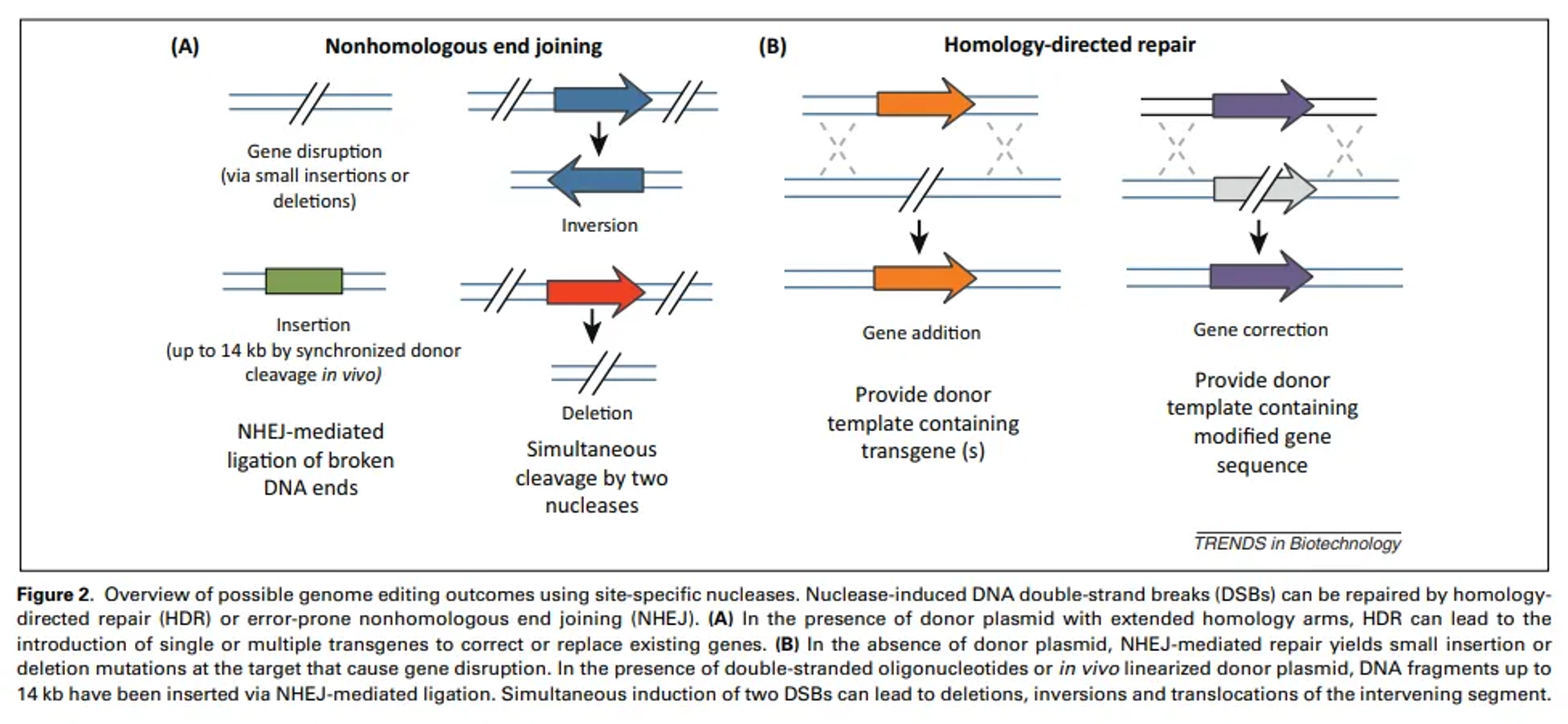
Homology-Directed Repair (HDR)
Homology-Directed Repair (HDR) is a precise DNA repair mechanism that cells use to fix breaks in double-stranded DNA by using a similar or identical DNA sequence as a template. This process ensures accurate repair, preserving the genetic information.
Step-by-Step Working of HDR
HDR is a high-fidelity DNA repair mechanism that uses a homologous sequence as a template to accurately repair DSBs. This process ensures the genetic information is restored precisely.
1. Detection of DNA Break:
MRN Complex Activation:
- When a DSB occurs, the MRN complex (composed of Mre11, Rad50, and Nbs1) detects the break.
- The MRN complex binds to the DNA ends and recruits the ATM (Ataxia-Telangiectasia Mutated) kinase, which phosphorylates various substrates to activate the DNA damage response.
2. End Resection:
Initial Resection:
- The MRN complex, along with CtIP (CtBP-interacting protein), initiates the resection of the 5' ends of the break, creating short 3' single-stranded DNA (ssDNA) overhangs.
Extended Resection:
- The initial resection is extended by nucleases such as Exo1 and Dna2, in cooperation with helicases like BLM or WRN. This process generates longer 3' ssDNA tails.
3. Coating of ssDNA:
RPA Binding:
- The ssDNA overhangs are rapidly coated with Replication Protein A (RPA). RPA stabilises the ssDNA and prevents the formation of secondary structures.
Rad51 Loading:
- BRCA1, BRCA2, and other mediator proteins help displace RPA and load Rad51 recombinase onto the ssDNA. Rad51 forms a nucleoprotein filament that is essential for the search for homology and strand invasion.
4. Homology Search and Strand Invasion:
Homology Search:
- The Rad51-coated ssDNA filament searches for a homologous sequence in a sister chromatid (or an exogenous donor template if provided). This search is facilitated by the high processivity of Rad51.
Strand Invasion:
- Once a homologous sequence is found, the ssDNA filament invades the double-stranded homologous DNA, forming a displacement loop (D-loop). The invading strand pairs with the complementary strand of the homologous DNA.
5. DNA Synthesis:
Primer Extension:
- DNA polymerase extends the 3' end of the invading strand using the homologous sequence as a template. This extension synthesises new DNA, accurately copying the sequence from the template.
D-loop Migration:
- As DNA synthesis progresses, the D-loop migrates, allowing further synthesis and stabilisation of the repaired DNA.
6. Resolution of Joint Molecules:
Displacement and Annealing:
- The newly synthesised strand is eventually displaced from the homologous template and pairs with the other 3' ssDNA tail at the break site.
- This step involves annealing the complementary sequences at the break site.
Second-End Capture:
- The second end of the break is captured and annealed with the newly synthesised DNA. This step ensures that both ends of the DSB are accurately repaired.
7. Ligation:
Gap Filling and Ligation: DNA polymerases fill any remaining gaps in the DNA. The newly synthesised DNA is ligated to the original DNA by DNA ligases, completing the repair process.
Formation of Holliday Junctions: In some cases, double Holliday junctions are formed. These junctions are resolved by specific resolvases, ensuring the proper segregation of the repaired DNA molecules.
Importance and Characteristics of HDR:
High Fidelity: HDR is highly accurate because it uses a homologous sequence as a template, ensuring that the repaired DNA is identical to the original.
Cell Cycle Dependence: HDR is most active during the S and G2 phases of the cell cycle when sister chromatids are available as templates.
Applications in Gene Editing: HDR is exploited in gene editing technologies, such as CRISPR-Cas9, to introduce precise changes in the genome by providing an exogenous DNA template with the desired sequence.