Sunday, October 6, 2024
sgRNA-DNA sequence encoding: Paper Analysis and Research Scope

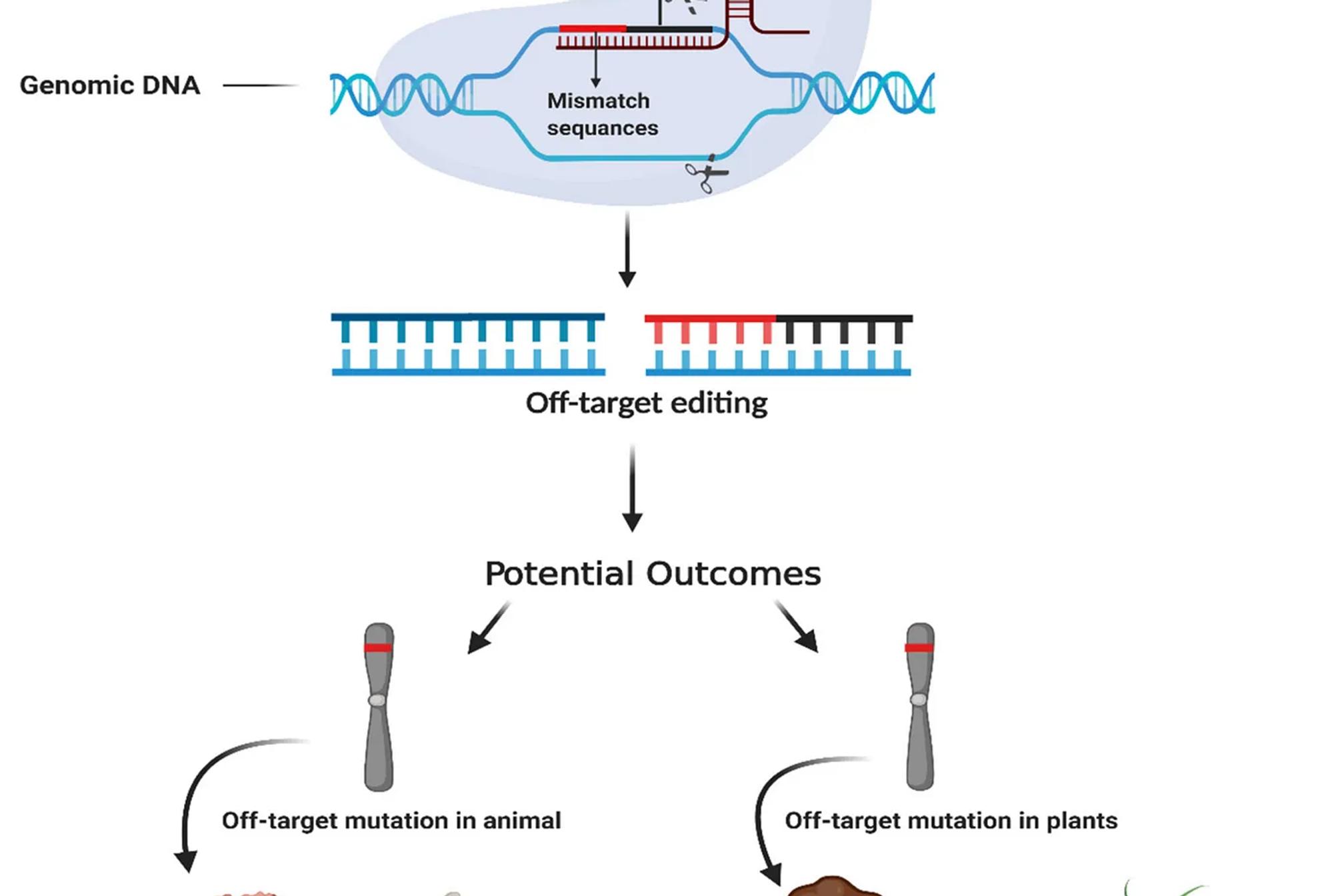
Overview
The CRISPR-Cas9 gene editing technique has revolutionized biotechnology by enabling precise DNA modifications. However, one of its significant challenges is off-target effects, which occur when the Cas9 enzyme unintentionally edits similar but unintended locations in the genome. These off-target mutations can have detrimental consequences, especially in therapeutic applications. To address this problem, the research paper presents a novel deep learning-based approach to improve off-target prediction accuracy in CRISPR-Cas9, focusing on the interaction between sgRNA (single guide RNA) sequences and DNA.
The research introduces an innovative encoding method to better represent the interactions between sgRNA and DNA sequences. This technique improves prediction accuracy when compared to traditional methods. The paper compares different deep learning models, such as Convolutional Neural Networks (CNNs), Recurrent Neural Networks (RNNs), and Feedforward Neural Networks (FNNs), and highlights significant improvements in off-target prediction.
Framework and Architecture
- Data Preparation and Sequence Encoding The input data consists of pairs of sgRNA and DNA sequences. In the CRISPR-Cas9 system, sgRNA guides the Cas9 enzyme to a target DNA sequence where it performs a cut. However, off-target effects happen when Cas9 accidentally binds to a DNA sequence resembling the intended target.
- Traditional Encoding: Conventional encoding methods combine both sgRNA and DNA into a single 4×L matrix. This method risks information loss, as it merges the representation of sgRNA and DNA prematurely.
- Proposed 8×L Encoding: The novel encoding scheme addresses this issue by encoding the sgRNA and DNA sequences separately in an 8×L matrix. Each nucleotide (A, T, C, G) is represented using a one-hot encoding vector. The sgRNA sequence occupies the first four rows of the matrix, while the DNA sequence takes the next four rows. This allows the model to learn intricate interactions between the sequences more effectively, retaining all information without merging
Model Architecture
The authors explored multiple deep learning architectures to predict off-target effects based on the encoded sequence data.
Convolutional Neural Networks (CNNs):
- Convolutional Layers: CNNs are used to extract local patterns in the sequence data. These patterns reveal key interactions between sgRNA and DNA, which may contribute to off-target effects.
- Activation Function: After convolutional filters are applied, ReLU activation introduces non-linearity, allowing the network to learn complex patterns.
- Pooling Layers: MaxPooling is applied to reduce the dimensionality of feature maps, improving efficiency and preventing overfitting.
- Fully Connected Layers: The features extracted by the CNN are flattened and passed to fully connected layers, which combine the patterns to make the final prediction.
- Output Layer: A final neuron with a sigmoid activation function outputs the probability that an off-target effect will occur.
Recurrent Neural Networks (RNNs):
- Bidirectional Long Short-Term Memory (BiLSTM) layers allow the model to learn sequence dependencies, crucial for understanding how different nucleotides in the sgRNA and DNA sequences interact.
Memory Mechanism: LSTMs capture the long-term dependencies within sequences, which are vital for determining how nucleotides far apart in the sequence might influence off-target activity.
Feedforward Neural Networks (FNNs):
- In FNNs, the encoded matrix is flattened into a vector and directly fed into dense layers. While it’s simpler than CNNs or RNNs, FNNs still model interactions between sgRNA and DNA sequences, but they lack the spatial awareness of CNNs.
Training Process
- Loss Function: A binary cross-entropy loss function is used to evaluate the difference between predicted and actual off-target outcomes.
- Optimization: The Adam optimizer is employed, adjusting learning rates adaptively for faster convergence.
- Regularization: Techniques like dropout are implemented to prevent overfitting and improve model generalization on unseen data.
Evaluation Metrics
- The models are evaluated using the AUC-ROC (Area Under the Receiver Operating Characteristic) curve, a metric that measures the model’s ability to distinguish between true positives and false positives.
- The best-performing models typically involve CNNs due to their superior ability to extract local patterns in sequence data, resulting in more accurate off-target predictions.
Datasets Used
GUIDE-seq Dataset:
- Provides validated data on double-strand breaks (DSBs) caused by CRISPR-Cas9. Although small in size, it offers highly reliable data for training and testing models.
CRISPOR Dataset:
- A larger dataset, consisting of over 26,000 sgRNA-DNA pairs. It includes both validated and computationally predicted off-target sites, making it an extensive resource for training deep learning models.
Scope for Future Research and Improvements
While this study shows promising results in improving off-target predictions, there is ample opportunity for further research and optimization. The following improvements could be explored:
- CNN-ConvLSTM Model: Combining CNN for local feature extraction and ConvLSTM for temporal relationships could improve the accuracy of predictions by accounting for both local and sequential dependencies in the data.
- VGG16 for Experiments: Leveraging deeper models like VGG16, known for their powerful feature extraction, could provide more refined predictions, though this may be computationally expensive.
- Combined Embeddings: A potential improvement involves using combined embeddings for sgRNA and DNA sequences, possibly with k-mer encoding and GloVe embeddings. This can capture the context in which nucleotides appear in sequences, similar to natural language processing (NLP) models.
- Attention-based BiLSTM: Incorporating an attention mechanism into the BiLSTM framework would allow the model to focus on the most relevant parts of the sequence when predicting off-target effects.
- Transformer or BERT-based Models: Recently, Transformer models, such as BERT or GPT, have shown exceptional performance in learning complex patterns in sequential data. Applying these NLP models to CRISPR sequence prediction could further enhance prediction accuracy.
Conclusion
The research presents an advanced deep learning approach to predicting off-target effects in CRISPR-Cas9 gene editing by introducing a novel 8×L sequence encoding scheme. The use of CNNs, FNNs, and RNNs, combined with innovative encoding, significantly improves prediction accuracy compared to traditional methods. While the current results are promising, there is immense scope for further advancements in this field by exploring more sophisticated models and embedding techniques. These improvements have the potential to significantly reduce the off-target effects of CRISPR-Cas9, pushing the boundaries of precision gene editing and its clinical applications.