Sunday, October 6, 2024
Apindel: Detailed Literature Review and Research Scope

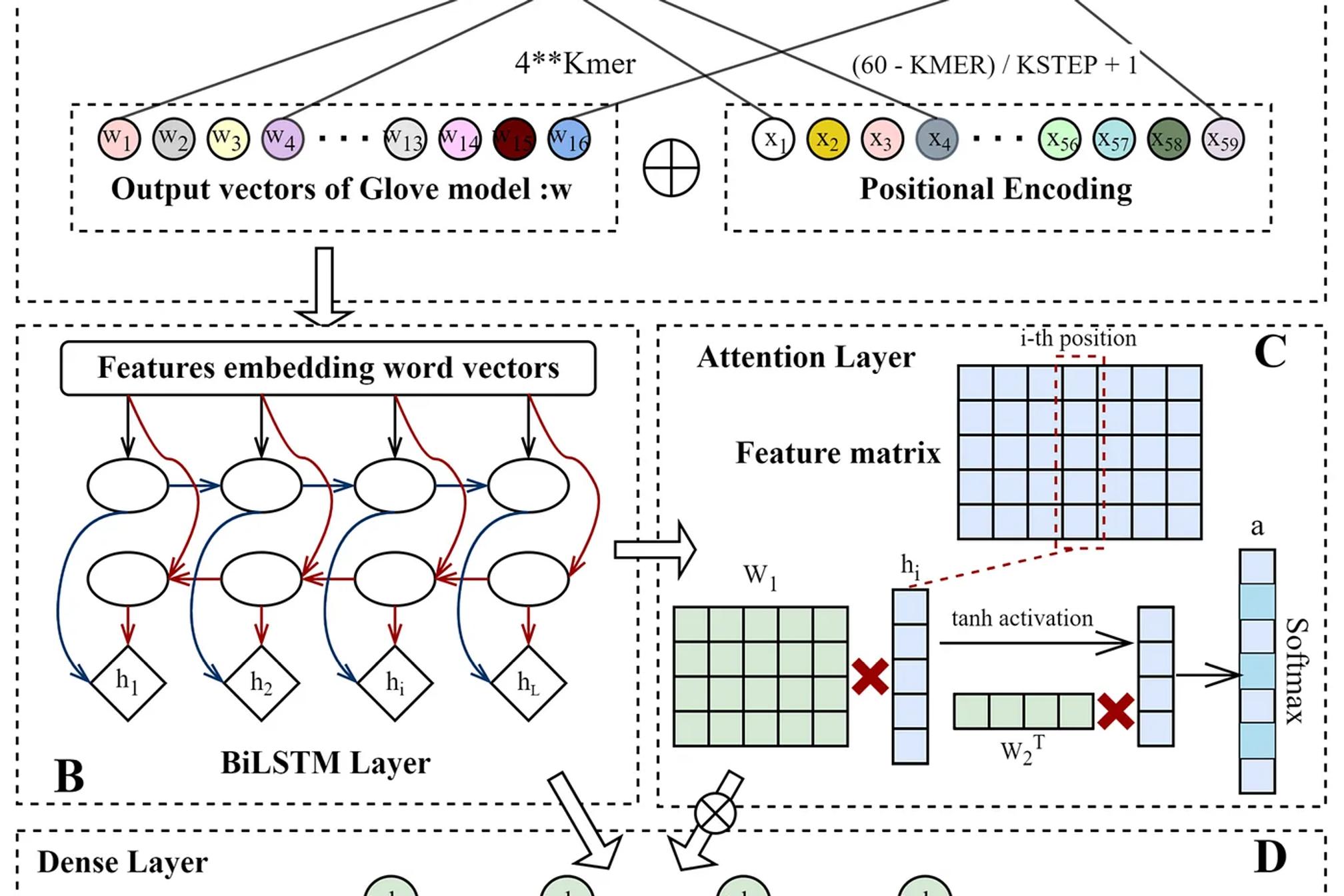
The paper titled “Predicting CRISPR/Cas9 Repair Outcomes by Attention-Based Deep Learning Framework” introduces the Apindel model, a deep learning architecture designed to predict DNA repair outcomes post-CRISPR/Cas9 gene editing. This literature review delves into the methodologies, dataset details, and potential improvements.
1. Dataset Overview
Two main datasets, Lindel and SPROUT, are employed for training and evaluating the Apindel model. Both datasets contain information on DNA sequences and CRISPR-induced repair outcomes such as insertions and deletions.
- Lindel Dataset: Provides information on CRISPR/Cas9 cleavage outcomes and focuses on indel mutations, offering a substantial collection of sequences with documented repair outcomes.
- SPROUT Dataset: Complements the Lindel dataset by offering a broader view of CRISPR/Cas9 repair events, including more complex mutations like frameshifts. This diversity enhances model robustness.
Key Information Extracted:
- Sequences: Nucleotide sequences surrounding the CRISPR/Cas9 cut site.
- Repair Outcomes: Labels specifying the nature of the repair event (e.g., insertion, deletion).
- Positional Data: Location information that contextualizes how nucleotide position relative to the cleavage site affects repair outcomes.
2. Model Architecture and Workflow
The Apindel model follows a structured approach to process the DNA sequences and predict repair outcomes:
Step 1: Input Sequence Representation
- k-mer Embedding: The model breaks down sequences into k-mers, where k represents the length of the subsequence. This technique helps capture local patterns within the DNA.
- GloVe Embedding: Inspired by natural language processing, GloVe embeddings map k-mers into dense vectors, enabling the model to learn semantic similarities between different subsequences.
- Positional Encoding: Embeds the nucleotide’s position in the sequence to highlight the importance of relative location, particularly around the CRISPR cut site.
Step 2: BiLSTM Layer
- Bidirectional LSTM (BiLSTM): A recurrent neural network layer processes sequences in both forward and backward directions, capturing long-range dependencies in DNA. This layer learns how specific nucleotides interact with others and contribute to repair outcomes.
Step 3: Attention Mechanism
- Attention Layer: After the BiLSTM layer, an attention mechanism assigns weights to various parts of the sequence, indicating which regions are most influential in predicting repair outcomes.
- High-Attention Regions: Typically, regions near the cleavage site receive the highest attention scores, as these are critical in the DNA repair process.
Step 4: Output Layer
- Prediction Layer: The model’s final layer outputs the likelihood of different repair events (e.g., insertion, deletion), using the attention-weighted sequence information.
3. Training and Evaluation
Training Process: The model is trained on sequences from the Lindel and SPROUT datasets, using a categorical cross-entropy loss function to optimize the model’s accuracy in predicting the repair outcomes.
Model Evaluation: Performance is assessed using a separate test set, with standard metrics like accuracy, precision, recall, and F1-score. The Apindel model’s results are then compared to other models such as inDelphi, CROTON, and SPROUT.
4. Results and Insights
Superior Performance: Apindel outperforms competing models, especially in predicting common events like single base pair insertions and deletions.
Challenges: Predicting more complex mutations, such as frameshifts, remains an area for improvement. The model struggles to accurately forecast these rarer and more intricate repair events.
5. Future Scope and Improvements
There are several ways the model can be further enhanced:
Improvements in Embeddings:
- Replacement of GloVe Embeddings: Using Transformer-based or BERT-based embeddings, which excel in capturing context, could significantly improve feature representation.
- GPT-based Embeddings: Pretrained models on DNA sequences can help in richer feature extraction by leveraging sequence context more effectively.
Enhancing Model Architecture:
- Replacing BiLSTM with Transformer: Adopting transformer-based architectures could enhance sequence modeling by capturing longer dependencies more effectively.
- BERT or GPT for DNA Sequence Modeling: Applying pretrained BERT or GPT models, fine-tuned on DNA sequence datasets, can extract more meaningful representations of the DNA sequences.
Improving Complex Mutation Predictions:
- Advanced NLP Models: Pretrained models from the NLP domain could provide better insights into rare and complex mutation types, potentially improving the model’s ability to generalize across different repair outcomes.
Alternative Feature Extraction:
- One-Hot Encoding in CNN Models: Though less effective for capturing deep sequence interactions, using CNNs with one-hot encoding could offer a complementary method for simpler mutation prediction tasks.
Conclusion
The Apindel model represents a major advancement in the prediction of CRISPR/Cas9 repair outcomes by effectively combining k-mer embeddings, BiLSTM, and attention mechanisms. However, the model has potential for improvement, especially in handling more complex DNA repair events and incorporating more powerful sequence embedding techniques. By integrating advanced deep learning models and exploring broader biological features, future research could further improve the accuracy and versatility of DNA repair predictions in CRISPR editing.